Since UK Biobank began in 2003, our database has been enhanced by multiple amazing scientific projects. These projects are helping researchers to accelerate public health discoveries around the globe.
The world’s largest imaging project
Our imaging project has scanned 100,000 of our participants’ brains, hearts, abdomens and bones, with hundreds of millions of images captured and making it the world’s largest imaging project. UK Biobank’s global community of approved researchers are combining these imaging data with other information, like lifestyle habits and genes, to understand how the diseases of ageing develop.
The project is the result of a collaboration between the UK government-funded Medical Research Council (MRC), Wellcome, the British Heart Foundation (BHF), and Dementias Platform UK.
Detecting changes over time
Sixty thousand people will also return for a second set of scans as part of our repeat imaging project. Multiple scans allow researchers to understand how changes to people’s organs link to diseases such as dementia, heart disease, and cancer. Additional funding to re-scan the 60,000 participants is being provided by the Medical Research Council (MRC), the company Calico, and the philanthropic Chan Zuckerberg Initiative (CZI).
The imaging project has already been used to look at the structure of the heart in more detail than ever, see how stress affects the brain, promote earlier detection of Parkinson’s and much more.
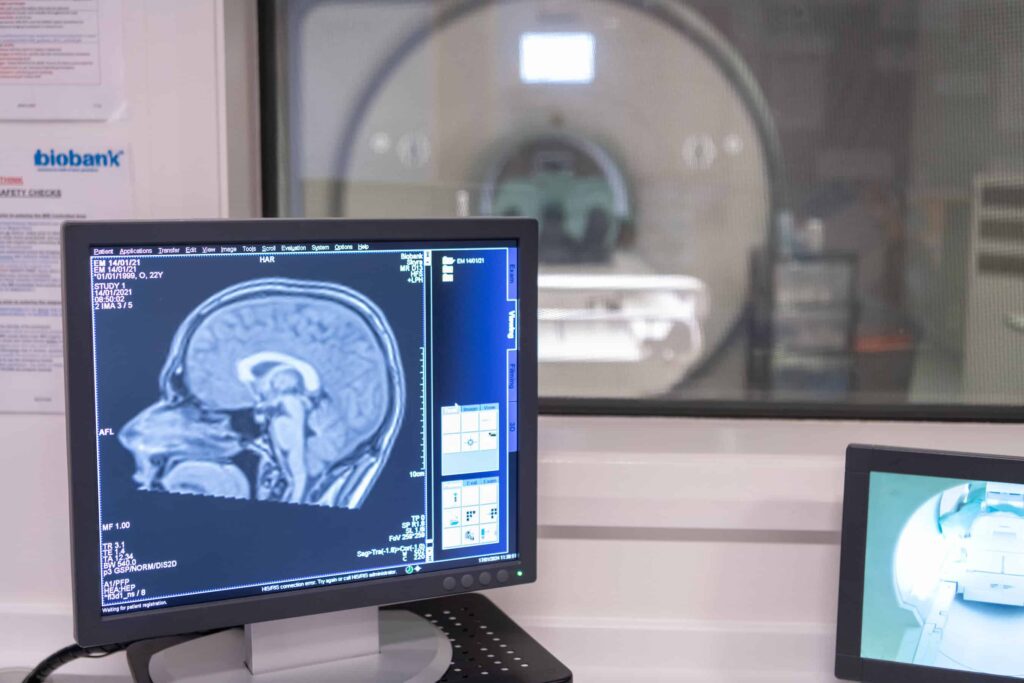

As UK Biobank reaches the record-breaking milestone of collecting 100,000 scans, learn more about the world’s largest imaging project and the impact that the imaging data is having on science.
Related content

Magnetic resonance images, bone-density scans, carotid artery ultrasound and more

Take part in our imaging project and help to transform research into diseases like dementia, heart disease, arthritis and diabetes.

In a remarkable achievement that is already impacting how we detect and diagnose disease, UK Biobank has completed the world’s largest whole body imaging project, scanning the brains, hearts, abdomens, blood vessels, bones and joints of 100,000 volunteers.

As UK Biobank’s record-breaking project crosses the finish line, researchers are working to reveal what the images can tell us about why we get ill as we age – and what to do about it.
The world’s largest set of whole exome sequences
UK Biobank also includes the world’s largest whole exome sequencing project, with data on over 470,000 participants available to approved researchers.
Whole exome sequencing measures the regions of the genome (about 2%) that are involved in coding for proteins, and is widely considered to be an important technique for identifying disease-causing or rare genetic variants.
Already, the exome sequencing data has resulted in novel genetic findings, such as for the prevention and treatment of obesity. A publicly accessible browsable resource, Genebass, has been made available to provide access to association summary statistics arising from these data.
The generation of whole exome sequencing data was made possible through collaboration and funding support from the UK Biobank Exome Sequencing Consortium (UKB-ESC). The consortium comprises of Regeneron Pharmaceuticals, AbbVie, Alnylam Pharmaceuticals, AstraZeneca, Biogen, Bristol Myers Squibb, Pfizer and Takeda.
Related content

Genotyping, exome and whole-genome information
One of the largest sets of whole genome sequences
UK Biobank includes data on all half a million participant’s whole genomes.
This data will help researchers to identify the genes that underpin common diseases. Understanding the genetic causes of these conditions will open up the discovery of new diagnostics, treatments and cures, and complement the wealth of data already available within UK Biobank.
This treasure trove of information has already allowed researchers to better understand how specific variations in a gene can have more than one impact. For example, a genetic variant linked to an increased life span was also found to increase the risk of head and neck cancer. Studies using these data have discovered genetic variants that are protective against obesity and type 2 diabetes, whilst other genetic variants may lead to a very high genetic risk for diseases such as heart disease, breast cancer and prostate cancer.
The Medical Research Council provided funding to UK Biobank in 2018 for a pilot project (the Vanguard) to perform whole-genome sequencing on 50,000 participants, which was undertaken by the Wellcome Sanger Institute, Cambridge.
A consortium of government, industry and charity then came together to fund whole genome sequencing of the remaining 450,000 participants. This project was funded by:
- UK Government’s research and innovation agency, UK Research and Innovation (UKRI), through the Industrial Strategy Challenge Fund
- Wellcome (a medical research charity)
- A consortium of industry partners: Amgen, AstraZeneca, GlaxoSmithKline and Johnson & Johnson.
A publicly available browsable resource, the UK Biobank Allele Frequency Browser, has been made available to provide access to variant allele frequencies arising from these data.
Related content

Genotyping, exome and whole-genome information
Data from 39,000 UK Biobank participants reveal how genetic variations that influence the size of brain structures also increase the risk for Parkinson’s and ADHD.
Faulty gene in body’s iron regulation found to play a role in polycythemia vera, a rare blood cancer.
World’s largest DNA analysis of people with depression reveals hundreds of inherited genetic differences, which could eventually help to match people with an antidepressant that works for them.
The world’s largest set of protein biomarkers
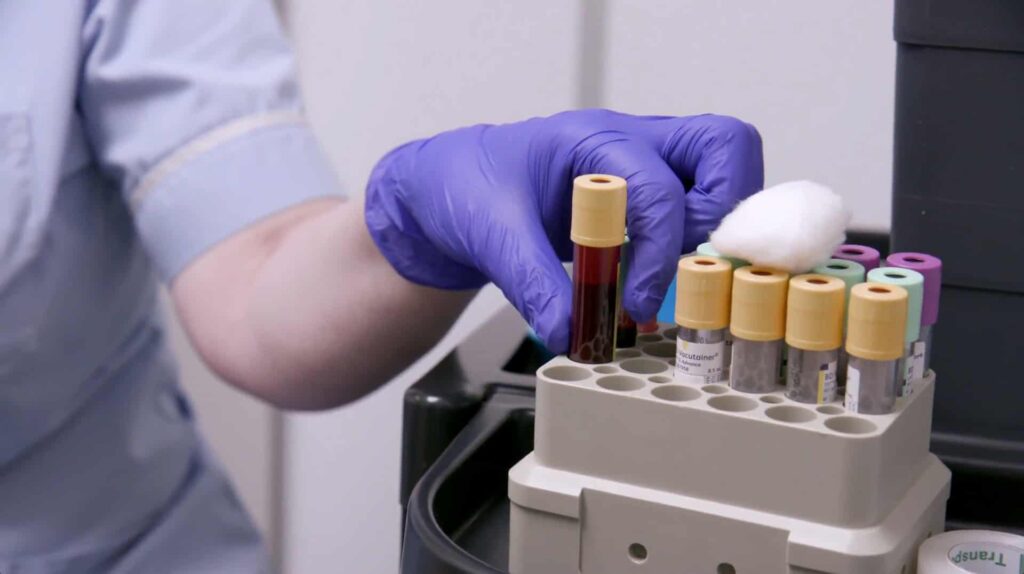
The UK Biobank dataset includes a bank of proteins that can be used to find markers for disease. This first-of-its-kind dataset is based on blood samples from over 54,000 UK Biobank participants. This research area, known as proteomics, provides a highly detailed picture of health. More than 14,000 specific links between proteins and health conditions have already been found.
This pilot work, funded by a consortium of 13 pharmaceutical companies, has already allowed scientists to identify four proteins linked to the onset of dementia and highlight proteins in the blood present more than seven years before a cancer diagnosis.
Making the protein dataset ten times bigger
We are expanding this set of protein markers by analysing 250,000 blood samples and 50,000 repeat samples taken from our altruistic volunteers.
The repeat samples allow researchers to compare protein levels up to 15 years later. Examining changes to protein levels as we age will shed light on how diseases develop, bringing new treatments and diagnostic tools closer.
This project aspires to measure up to 5,400 proteins in each of 600,000 samples, including those taken from half a million UK Biobank participants and 100,000 second samples taken from these volunteers up to 15 years later. A consortium of 14 pharmaceutical companies are funding the analysis of the first 300,000 samples, which will include initial samples from 250,000 UK Biobank volunteers and 50,000 second samples taken at follow-up assessments.
Related content

Proteins, metabolites, infectious disease markers and other biomarkers

Blood samples donated by more than 50,000 UK Biobank participants point to four biomolecules that can reveal developing dementia in apparently healthy people.
Our seminal COVID-19 study
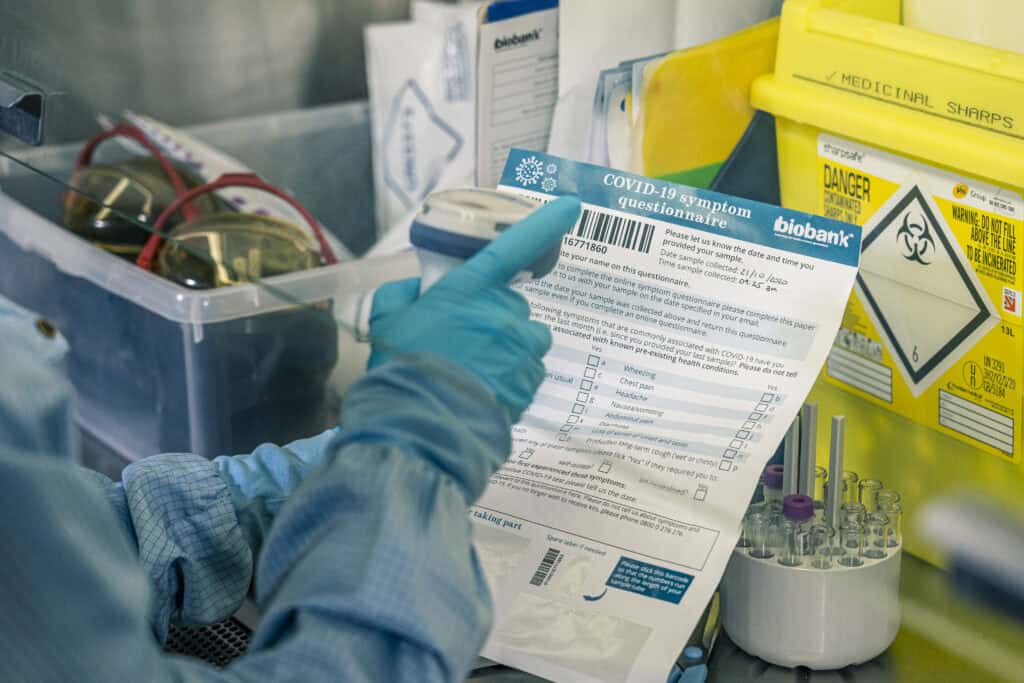
UK Biobank made a valuable contribution to tackling the global COVID-19 crisis. We made data available from electronic health records (diagnostic test data, death, hospitalisations and GP records) on a regular basis to researchers to better understand the genetic and lifestyle determinants of COVID-19.
In addition, 20,000 participants and their relatives provided regular blood samples over six months, and after one year, to help us understand the extent of infection and to provide insights about natural immunity to the virus. This project was funded by the UK Government’s Department of Health and Social Care.
We also sent out antibody test kits to all 500,000 participants to determine those previously infected with the virus to help us understand the longer-term health consequences of infection. Around 2,000 participants also attended a second imaging visit to understand the impact of the virus on internal organs.
These data continue to help researchers investigate the short-and long-term impacts of COVID-19 infection.
For example, repeat imaging data showed that even mild COVID-19 infections change the brain, with areas involved in smell processing getting smaller after infection.
Related content

Magnetic resonance images, bone-density scans, carotid artery ultrasound and more

Brain scans from almost 800 UK Biobank participants reveal damage in ‘smell centre’ – but it’s likely that the brain can heal itself.
UK Biobank, the UK’s major biomedical database and research resource, today reports the 6-month results of a major government-backed study of SARS-CoV-2 infection.
The UK Biobank Research Analysis Platform
The amount of data in UK Biobank and the number of researchers using it continues to grow. Currently, over 22,000 researchers based in more than 60 countries are using UK Biobank data. As a result, we needed a bespoke system, built to house the more than 30 petabytes of data collected.
The UK Biobank Research Analysis Platform (UKB-RAP) enables researchers to securely access our large-scale biomedical database in ‘the cloud’ from anywhere in the world. It has been designed and built by DNAnexus, and runs on Amazon Web Services (AWS) technology, to accommodate the vast and increasing scale of the UK Biobank resource.
Financial support to help offset the costs of analysing a huge amount of data is available to early career researchers and those from institutes in less wealthy countries courtesy of AWS. Additional funding for all other researchers is now also available courtesy of Wellcome.
The UKB-RAP allows researchers to use our growing database without the need for large amounts of computing power. The platform contains many tools needed for researchers to carry out their analyses and add to the growing number of publications powered by UK Biobank.
Related content

We made an important commitment to our participants when they joined the study. Find out how we uphold this commitment.
The UK Biobank Research Analysis Platform (UKB-RAP) enables researchers to securely access our large-scale biomedical database in ‘the cloud’ from anywhere in the world.
UK Biobank is a not-for-profit organisation dedicated to advancing global health research. To make our data as accessible as possible, we offer financial support through several funding programmes.
